Ensemble methods for survival analysis
Survival analysis is a collection of methods that analyse time-to-event data. The methods are used to estimate distribution of event times in a given population, and model its dependence on individual risk factors. The applications could be estimating a proportion of the initial population for which the event will happen by a certain point, or how the rate of event changes with time. It can also be employed to understand an impact of a risk factor, or make individual predictions on time-to-event. “Population” can be bacteria in a dish, group of people, group of financial companies, or app users; an event can be death, disease onset, cancer recovery, firm’s bankruptcy, or unsubscribing from an app.
We had health outcomes in mind (e.g. a disease onset) and aimed to develop user-friendly functions that enhance classical Cox proportionate hazards model with machine learning algorithms. The Cox model is one of the most popular model for health outcomes, and is essentially a general linear regression model. By embedding survival decision trees and survival random forest into the Cox model, we aimed to
-
boost predictive performance while estimating time-to-event by a given time point,
-
separate linear and non-linear relationships,
-
give insight into the data structure,
-
preserve algorithm’s interpretability.
All the code is in R, and can be found here
Methods
The methods’ description and initial results were presented at the talk at the NIHR Maudsley Biomedical Research Center Prediction Modelling Group meeting (September, 2021). More details are in the AIAI 2022 conference paper[1].
Graphical summary of the methods:
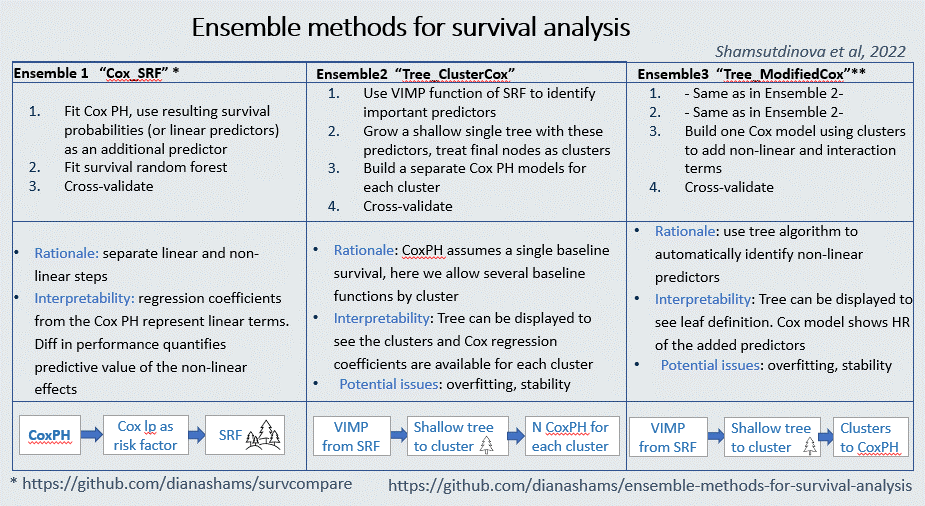
Ensemble 1 as an R package survcompare
The first ensemble method described in the paper [1] is now available as a stand alone R package. By comparing prediction performance of the Cox-PH and ensemble of Cox-PH and Survival Random Forest, it tests the presense of non-linearity and interactions in the data, and quantifies interpretability - accuracy trade off. That is, what gain in predictive accuracy one may get by employing a more complex model versus a baseline Cox-PH.
The package can be installed from source, and will soon be available on CRAN. More details and the code is available at survcompare page.
Links and references
[1] Shamsutdinova, D., Stamate, D., Roberts, A., & Stahl, D. (2022). Combining Cox Model and Tree-Based Algorithms to Boost Performance and Preserve Interpretability for Health Outcomes. In IFIP International Conference on Artificial Intelligence Applications and Innovations (pp. 170-181). Springer, Cham.
[2] Amunategui, M.: Data Exploration & Machine Learning, Hands-on. https://amunategui.github.io/survival-ensembles/index.html
[3] Marmerola, G.D.: Calibration of probabilities for tree-based models Guilherme’s Blog. https://gdmarmerola.github.io/probability-calibration/
[4] Su, X., Tsai, C.-L.: Tree-augmented Cox proportional hazards models. Biostatistics 6, 486–499 (2005)
[5] Breiman, L., Friedman, J.H., Olshen, R.A., Stone, C.J.: Classification and Regression Trees. Wadsworth & Brooks/Cole Advanced Books & Software, Monterey, CA (1984)
[6] Shimokawa, A., Kawasaki, Y., Miyaoka, E.: Comparison of splitting methods on survival tree. Int. J. Biostat. 11, 175–188 (2015)
[7] Ishwaran, H., Lauer, M.S., Blackstone, E.H., Lu, M.: randomForestSRC: Random Survival Forests Vignette (2021)
[8] Royston, P., Altman, D.G.: Regression using fractional polynomials of continuous covariates: parsimonious parametric modelling. Appl. Stat. 43, 429 (1994)
[9] Therneau, T., Atkinson, E.: An introduction to recursive partitioning using the RPART routines (2019)
[10] Ishwaran, H.: Variable importance in binary regression trees and forests. Electron. J. Stat. 1, 519–537 (2007)
Support or Contact
diana.shamsutdinova@kcl.ac.uk